|
Genome 5 genomic assemblies for D. alata, D. dumetorum, D. rotundata, D. tokoro, D. zingiberensis |
|
Transcriptome 14 de novo transcriptomic assemblies for D. polystachya, D. japonica, nipponica and more |

|
Plastome 41 complete chloroplast genomic assemblies and corresponding species phylogenic tree |

|
Variome 935 yam germplasms of D. rotundata and D. alata, together with 14 million SNPs and small InDels |

|
Pathway 479,819 genes of 19 yam species are organized in 145 signal pathways for exploring their functions |

|
Family Genes are classified into families and sub families based on Arabidopsis thaliana sequence similarity |

|
Collinearity six pair pariwise genome collinearity analysis for four yam genomes with chromosome-level assembly |

|
BLAST BLAST program is equipped for users to explore genes by sequence similarity |
|
Expression analysis 191 RNA-seq samples collected for differential gene expression and co-expression analysis |
|
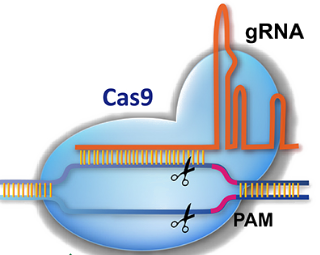
CRISPR sgRNAs CRISPR tool for designing CRISPR sgRNAs and estimate efficiency in genome editing |

|
Enrichment analysis Clusterfile is integrated to conduct functional enrichment analysis for specified gene set |
|
Phylogenetic placement Identification of kinship through construction of phylogenetic tree based on chloroplast genomes |