About
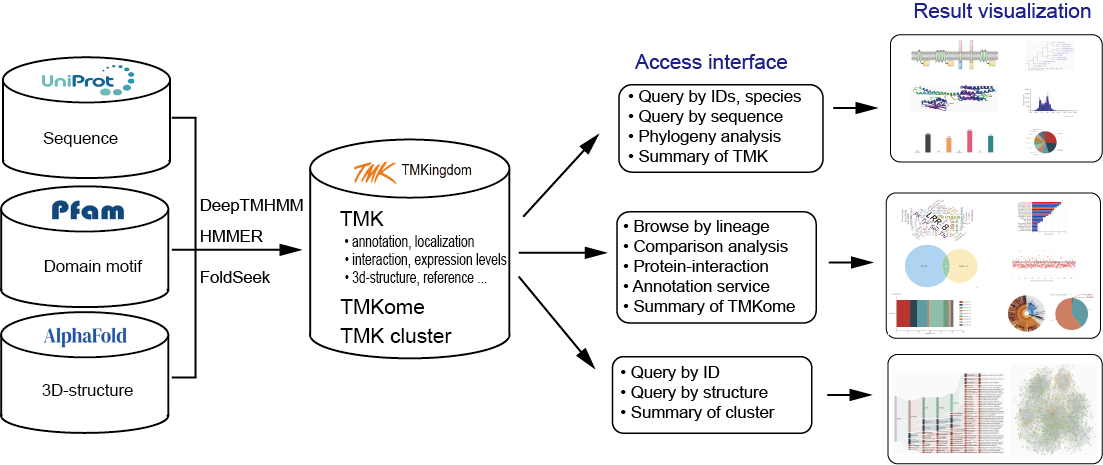
Transmembrane kinases (TMKs) are important cell surface mediators of signaling cascades that widely exist in the life kingdom and play key roles in diverse cellular responses to extracellular signals. TMKs from different kingdoms of life show distinct origins and functional diversity, which has not been fully explored yet. TMKingdom is a comprehensive data resource consisting of 1,415,870 TMKs from 33,047 species covering the whole life kingdom.
All data collected in the database are generally divided into three groups including TMKs, TMKome and TMK cluster. The friendly interfaces are developed for users conveniently to query or analyze each group of data. The retrieval result are visually displayed by charts or in interactive table.
Tutorial (Click blue titles to show details)
TMK interface
Search by ID or species
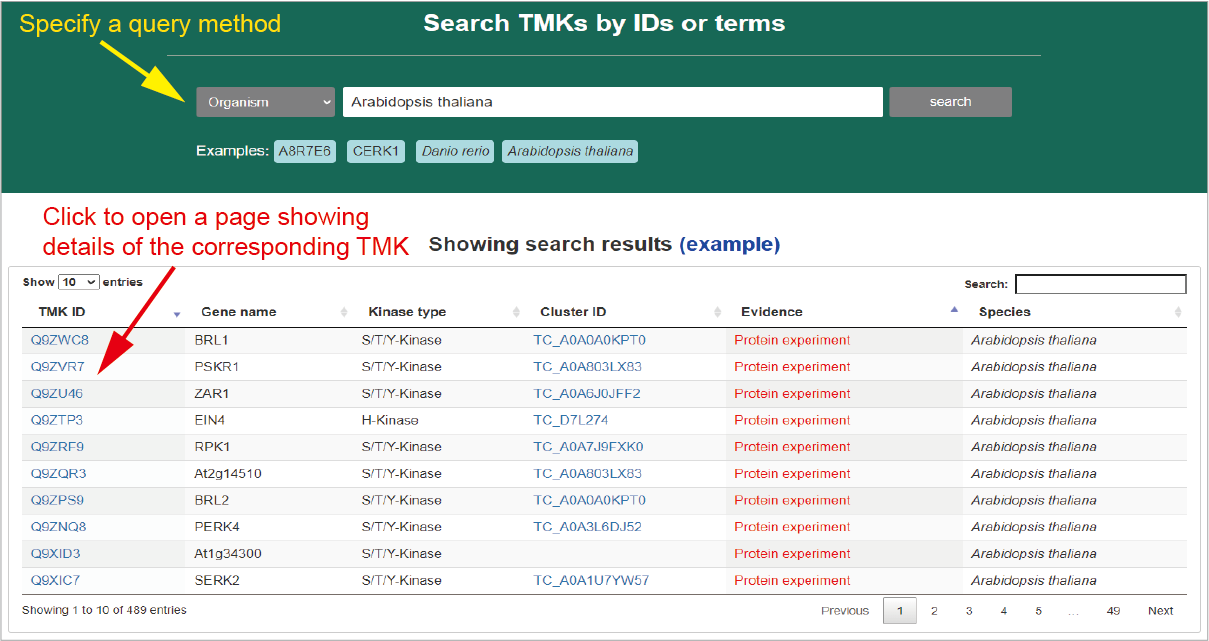
Through the TMK menu, users can open TMK search page to query TMKs. TMKs can be queried by IDs, protein name or species. After searching, results are listed in an interactive and sortable table below. By clicking on a TMK ID, a new page will be opened to show the details of the corresponding TMK.
Hide
Search by sequence
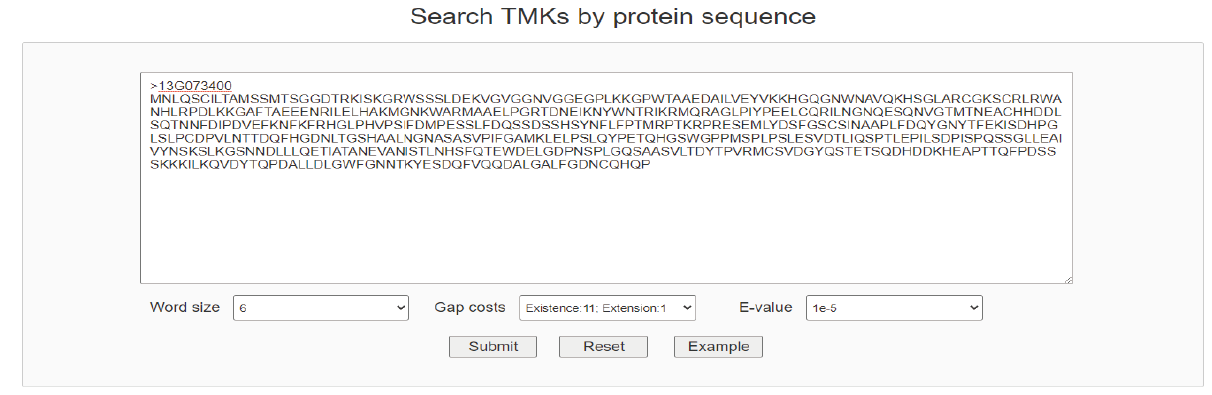
Through the TMK menu, users also can open a page to query TMKs by sequence similarity with blastp program. Furthermore, users can customize parameters to narrow the results. After searching, results are listed in an interactive and sortable table below.
Hide
TMK page content
The TMK page includes comprehensive information related to TMK, including basic information, annotation, expression, gene ontology, 3D-structure, orthologes and references.
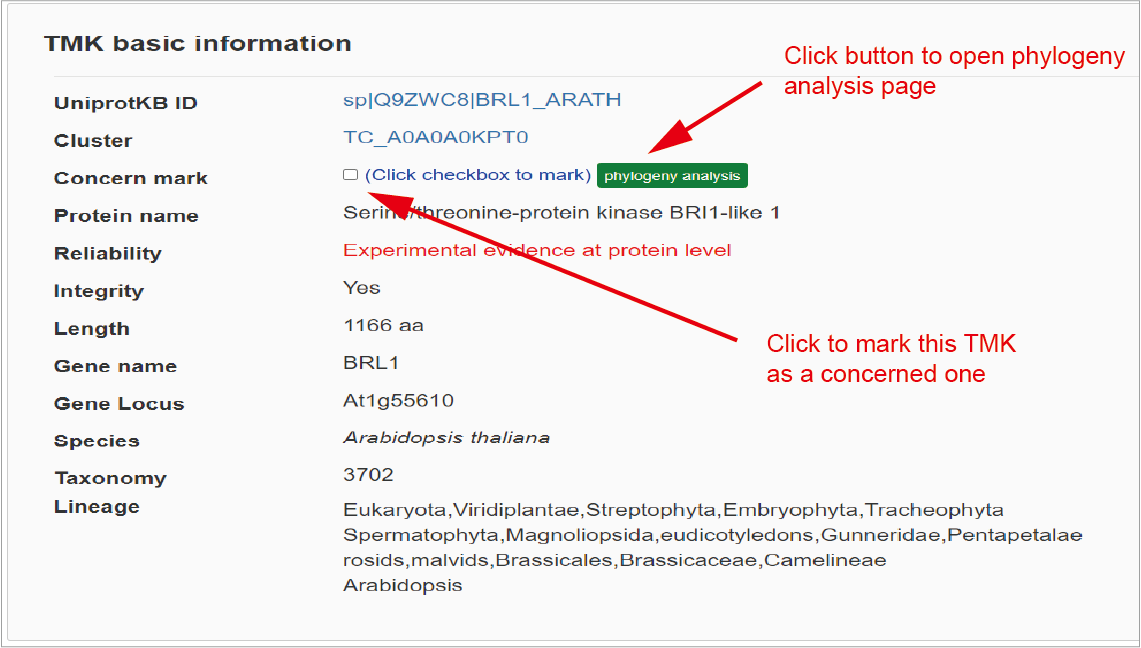
The first section of TMK page shows basic information including such as ID, cluster ID, protein name, reliability, length, gene name, origin species, lineage. If this TMK arouse interest of users, it can be marked for subsequent phylogeny analysis by clicking on the following button.Hide
The second section shows annotation information including kinase type, functional domains, comments, subcellar location and interaction. In addition to function domains annotated by pfam motif, improved domain annotation by 3D-structure similarity also is provided here.Hide
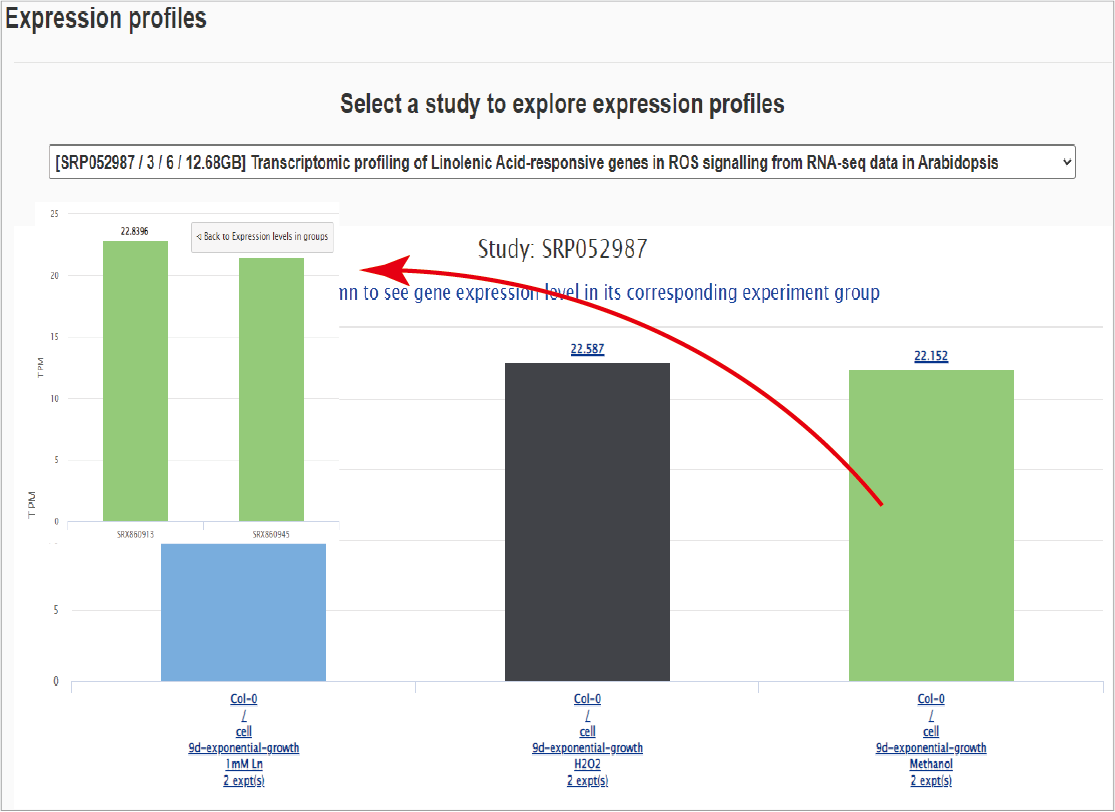
The third section shows corresponding gene express profiles in collected samples (if available). Click a sample group and the expression levels in each sample can be exhibited.Hide
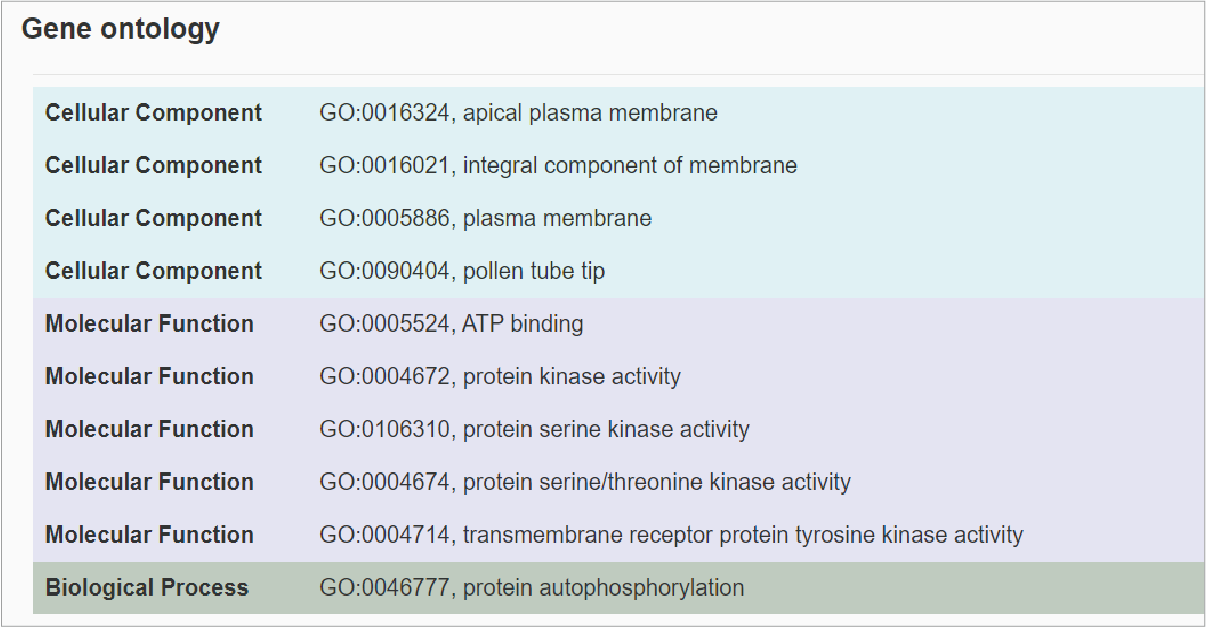
The forth section shows gene ontology description on three aspects including cellular component, molecular function and biological process.Hide
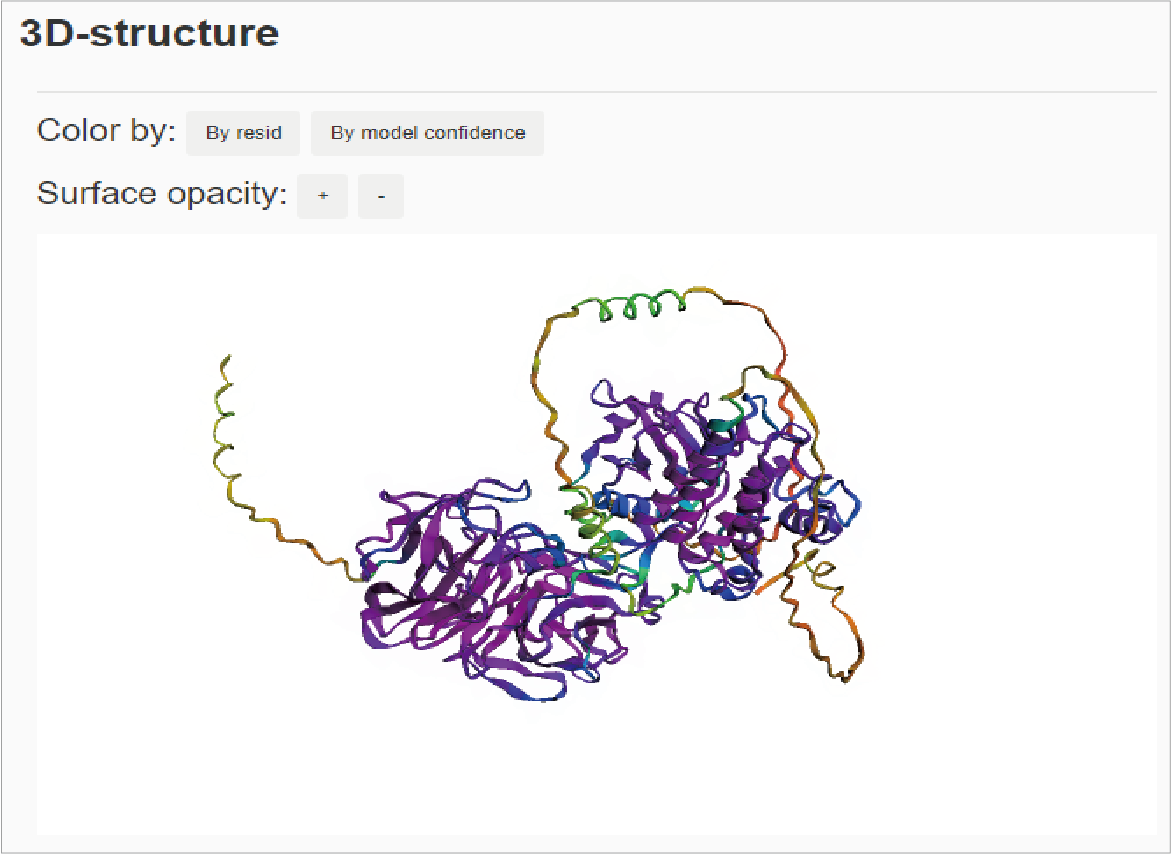
The fifth section shows 3D-structur of TMK. Users can adjust the structure graph by clicking on the buttons of color and surface opacity, and adjust the display angle by dragging the mouse.Hide
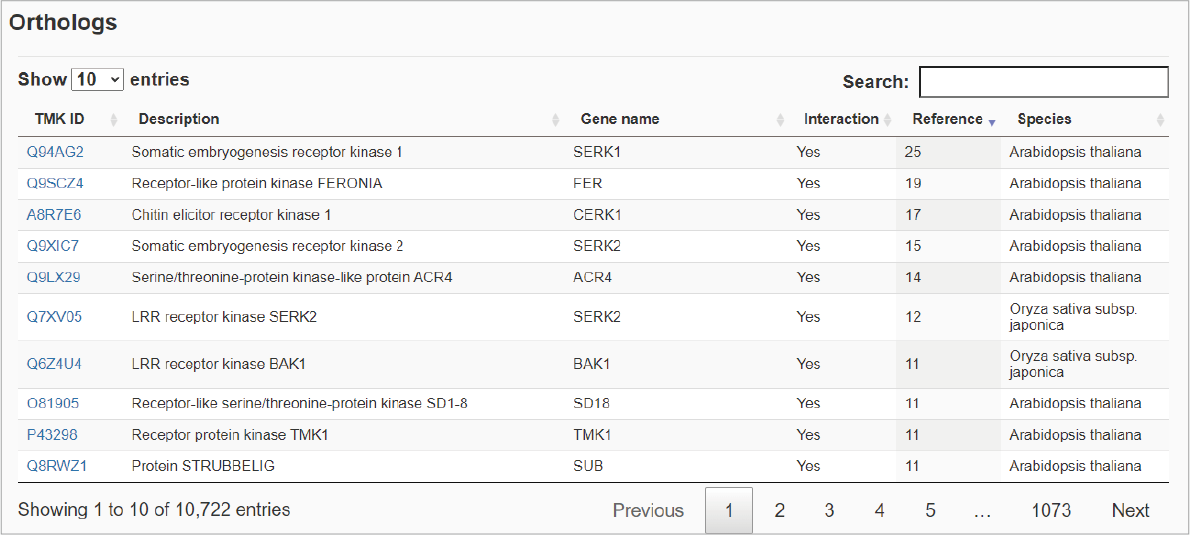
The sixth section shows orthologs with some basic information in an interactive table.Hide
The final section of TMK page shows references about this TMK.Hide
Phylogeny analysis
Through the TMK page or the Phylogeny analysis menu, users can open a page to investigate phylogeny of concerned TMKs. Users can specify any TMKs, then submit to conduct the analysis task. The phylogenic tree based on protein sequences will be exhibited and the global sequence alignments also can be further showed.
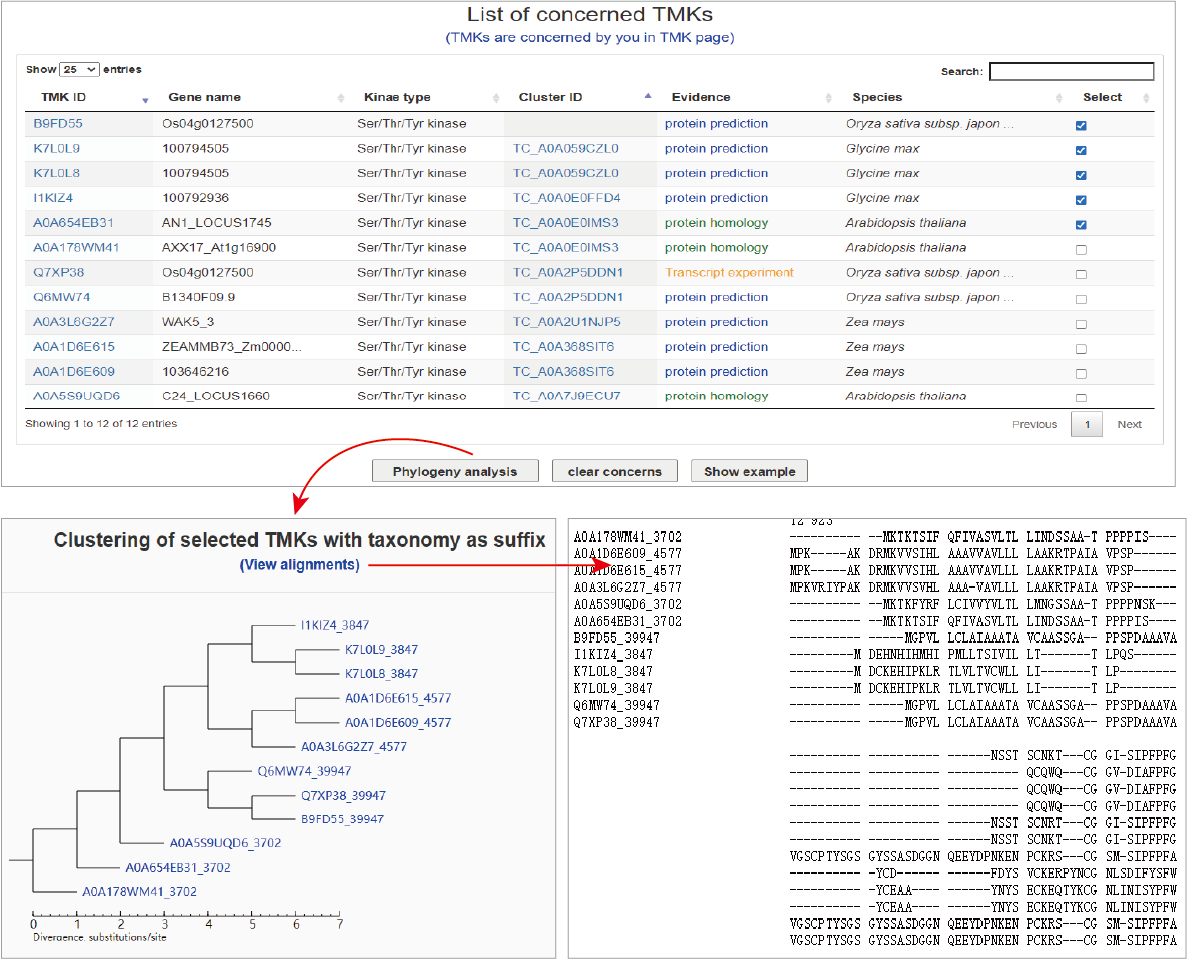
Hide
Summary of TMKs
The collected TMKs can be summarized in kinase type distribution, domain distribution and species distributions. The first section are pies showing the distribution of top 10 domains in each kinase type. Furthermore, users can specify a kinase type and domain to summarize distributions of species, sequence length and protein reliability. Finally, an interactive tabe shows the specified TMK numbers present in species.
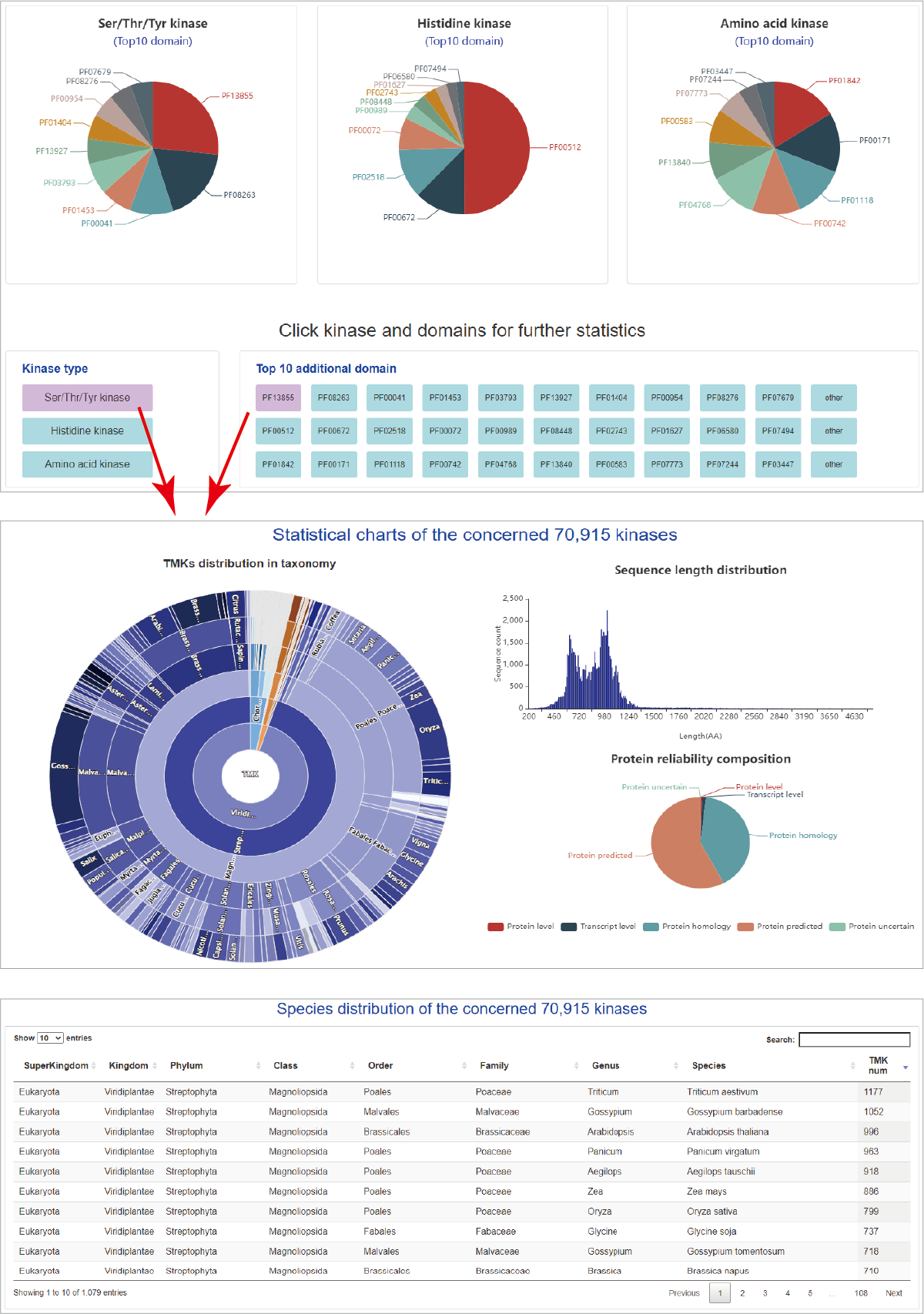
Hide
TMKome interface
Browse TMKomes by lineage
All species with TMKs collected in TMKingdom are organized as lineage tree. Users can specify a sub-lineage to browse TMKomes of concerned species. The concerned species are listed in an interactive table below with hyperlink to a TMKome page showing statistical charts of TMKs.
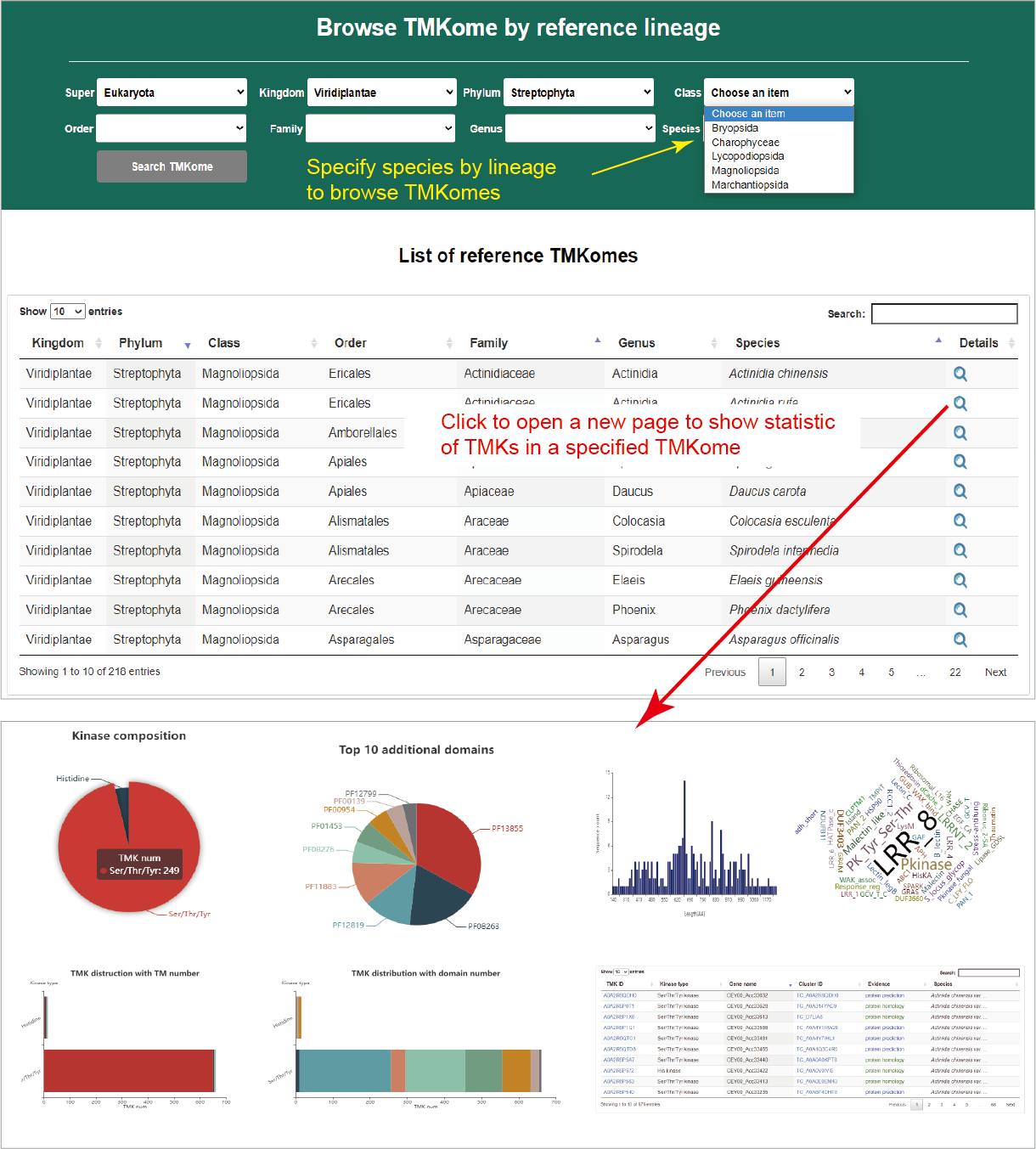
Hide
Comparison of TMKomes
Through the TMKome menu, users can open comarison analysis page. By specifying position in lineage tree, corresponding TMKomes will be listed in an interactive table with ratio buttons for setting comparison groups. After submit a comparison task, the analysis results will be exhibited below, including word cloud of domain showing frequency of domain, venn chart showing shared and differential domains and bar chart showing enriched domains for two groups. Finally, a table shows all domains in two TMKome groups.
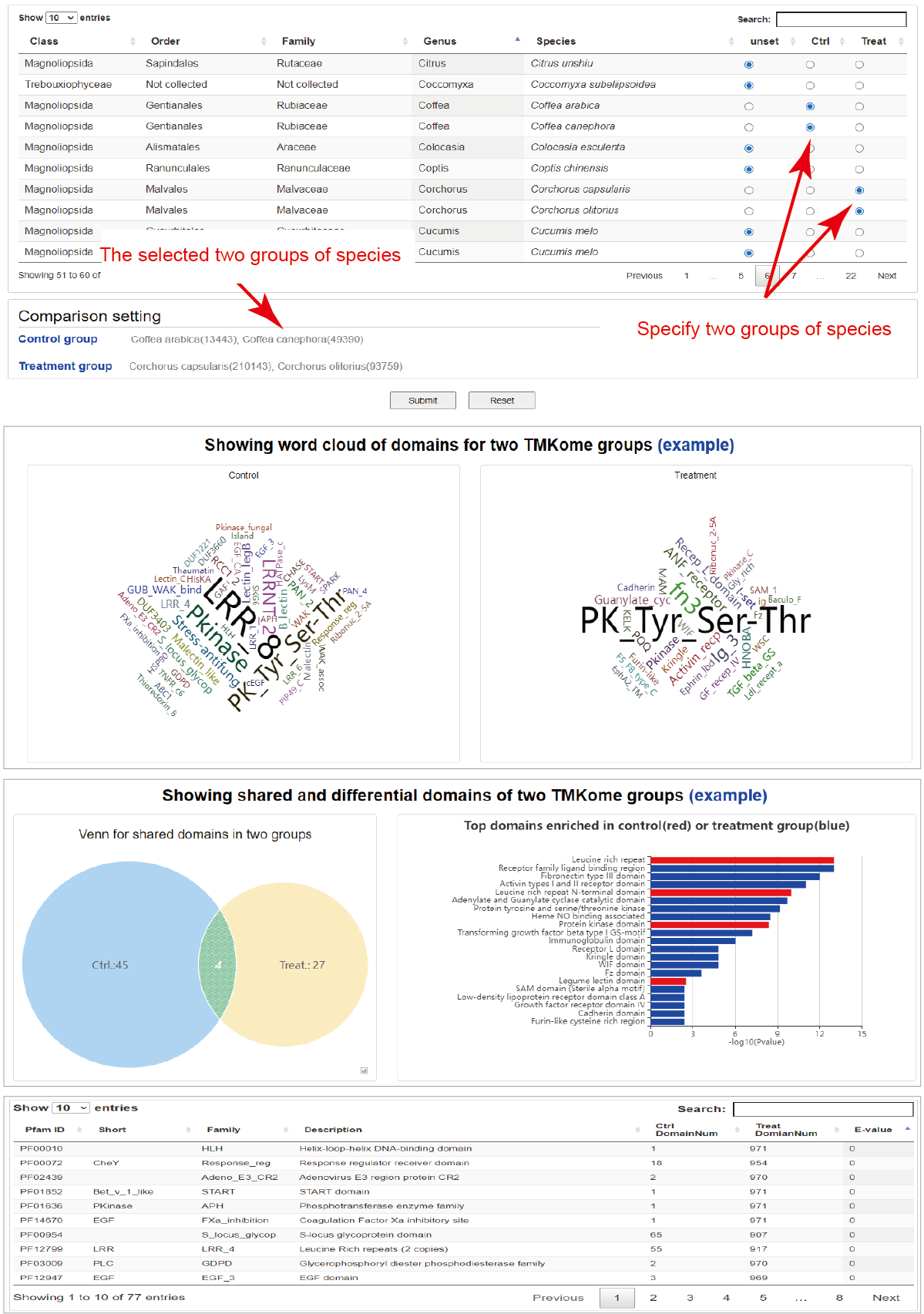
Hide
Protein-interaction prediction
TMKs tend to play roles by interaction with other ligand protein. Users can upload a protein 3D-structure to predict the possibility of interaction with TMKs in a specified TMKome. The higher PPI score, the greater the likehood of interaction. A PPI of >8 means interaction may occur. Prediction of interaction consumes a lot of computing resources. TMKingdom requires email to offline receive analysis results. In the result page, a scatter plot visually shows interaction score and all interactions are listed in a table with hyperlink to a page showing the details of hit TMKs.
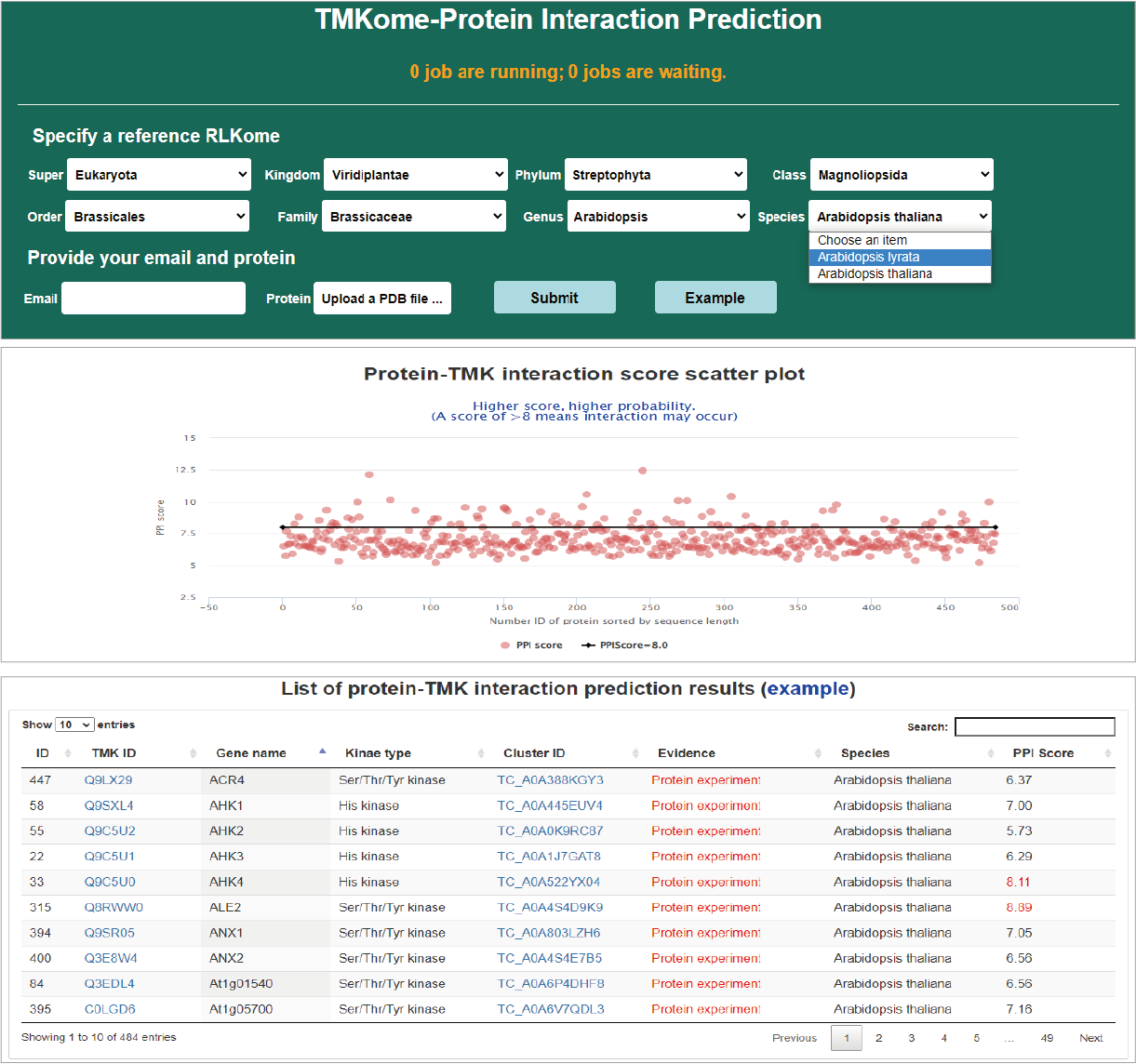
Hide
TMK annotation service
It's an important function that TMKingdom offers a service to annotate TMKs in a transcriptome or proteome. The annotation process consumes a lot of computing resource, so it also requires email to offline receive analysis result. In the result page, the identified TMKs are listed in a table with best hit TMKs collected in TMKingdom. The following are various charts summarizing TMK annotation.
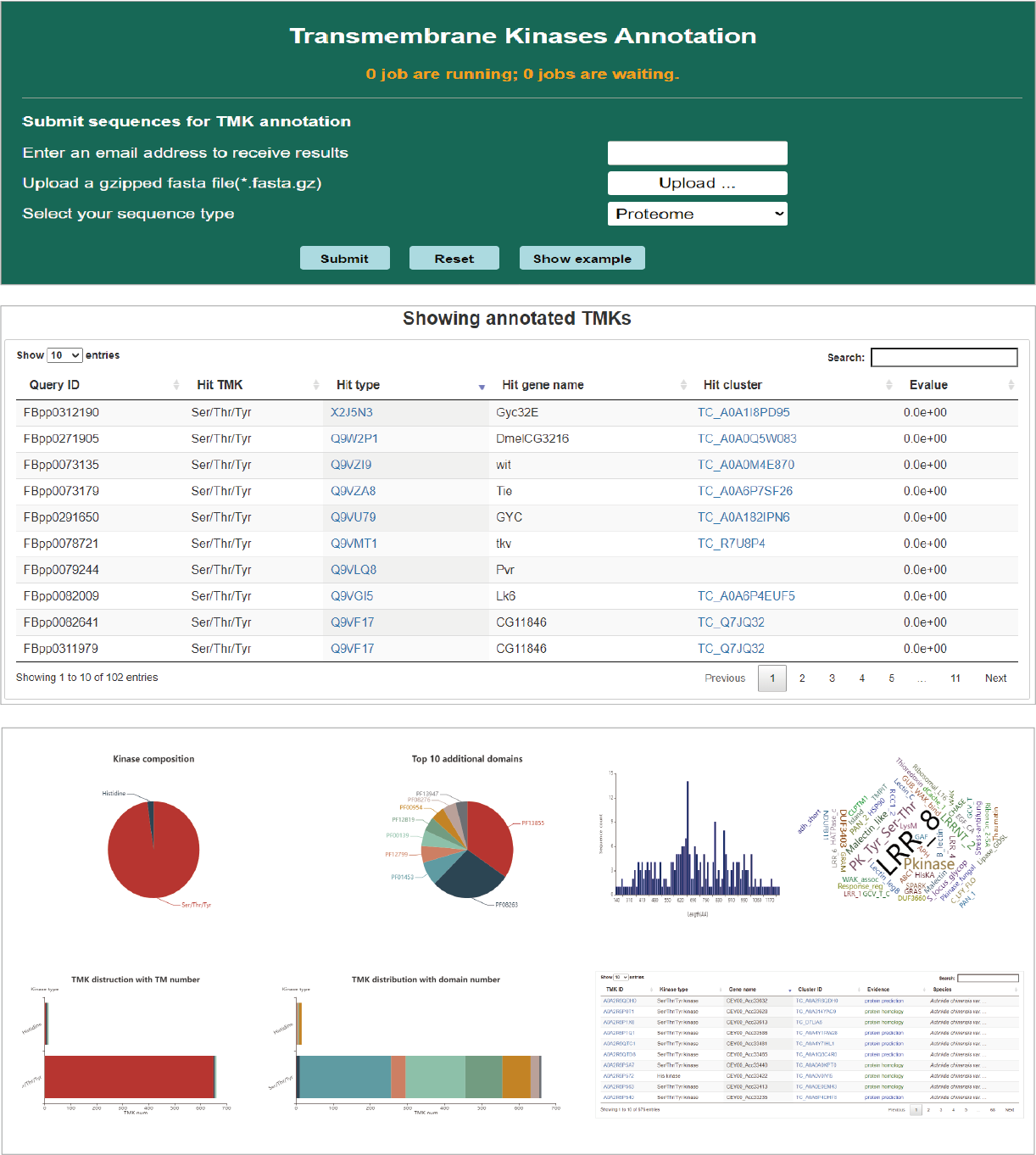
Hide
Summary of TMKomes
Users can summarize TMKs in any species by specifying a position in lineage. The several interactive charts are used to summarize TMKs in concerned species, such as species distribution, length distribution and reliability distribution. In addition, transmembrane number, kinase type, domain frequency are listed in an interactive table.
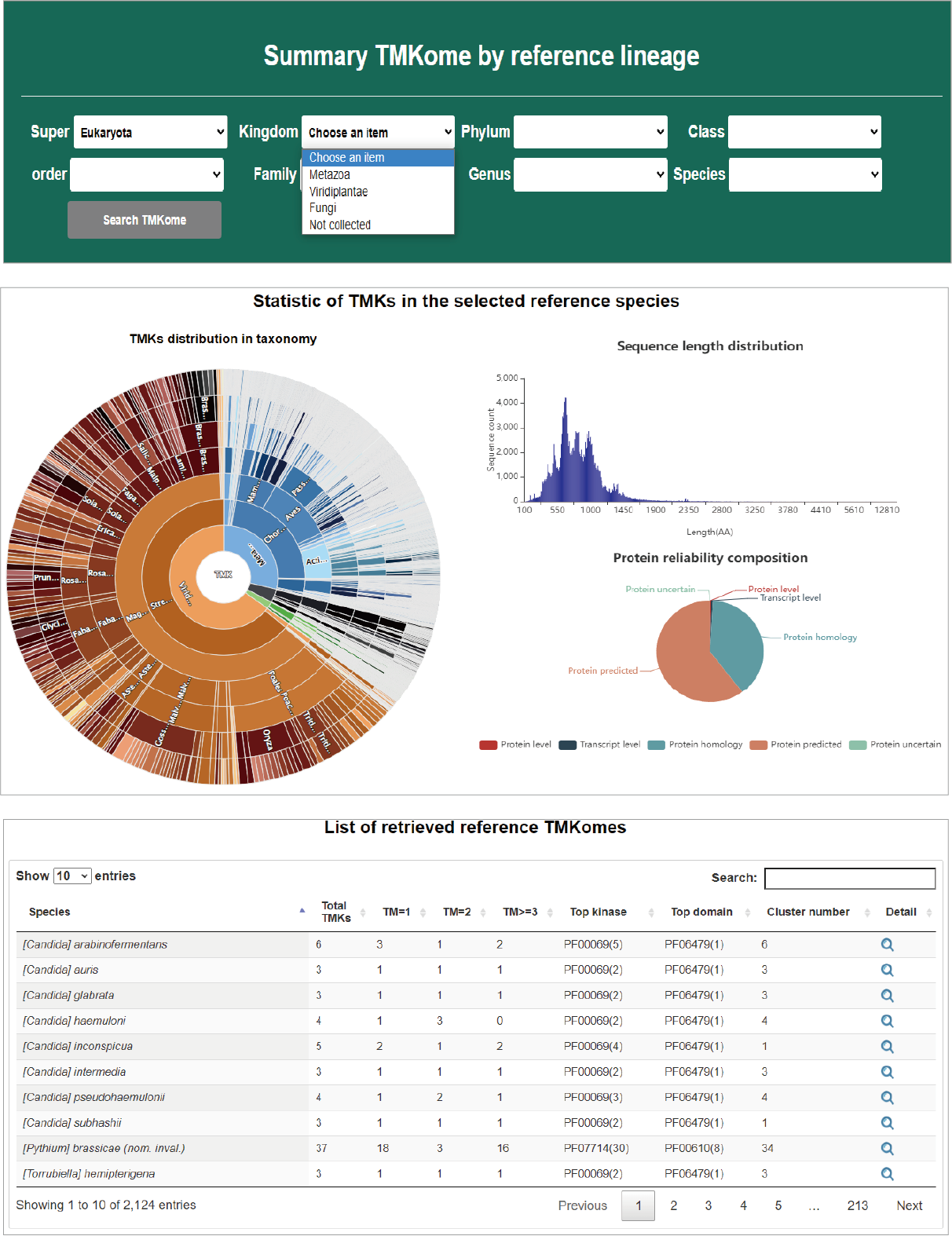
Hide
TMK cluster
Query by TMK/cluster ID
TMKingdom has an important hight that clustering TMKs based on 3D-structure covering the whole life kingdom. This highligth opens a new perspective for TMK function and evolution analyses. Users can search TMK cluster by TMK or cluster ID. First, the retrieved cluster will display its distribution in species lineage in the form of sankey. Second, a network shows the similarities among the members of a TMK cluster. Third, a pie chart exhibits the percentage of domains updated by 3D-structure similarity. Forth, a 3D-graph is used to exhibit the structure of a represent member. Finally, all TMK members are listed in an interactive table.
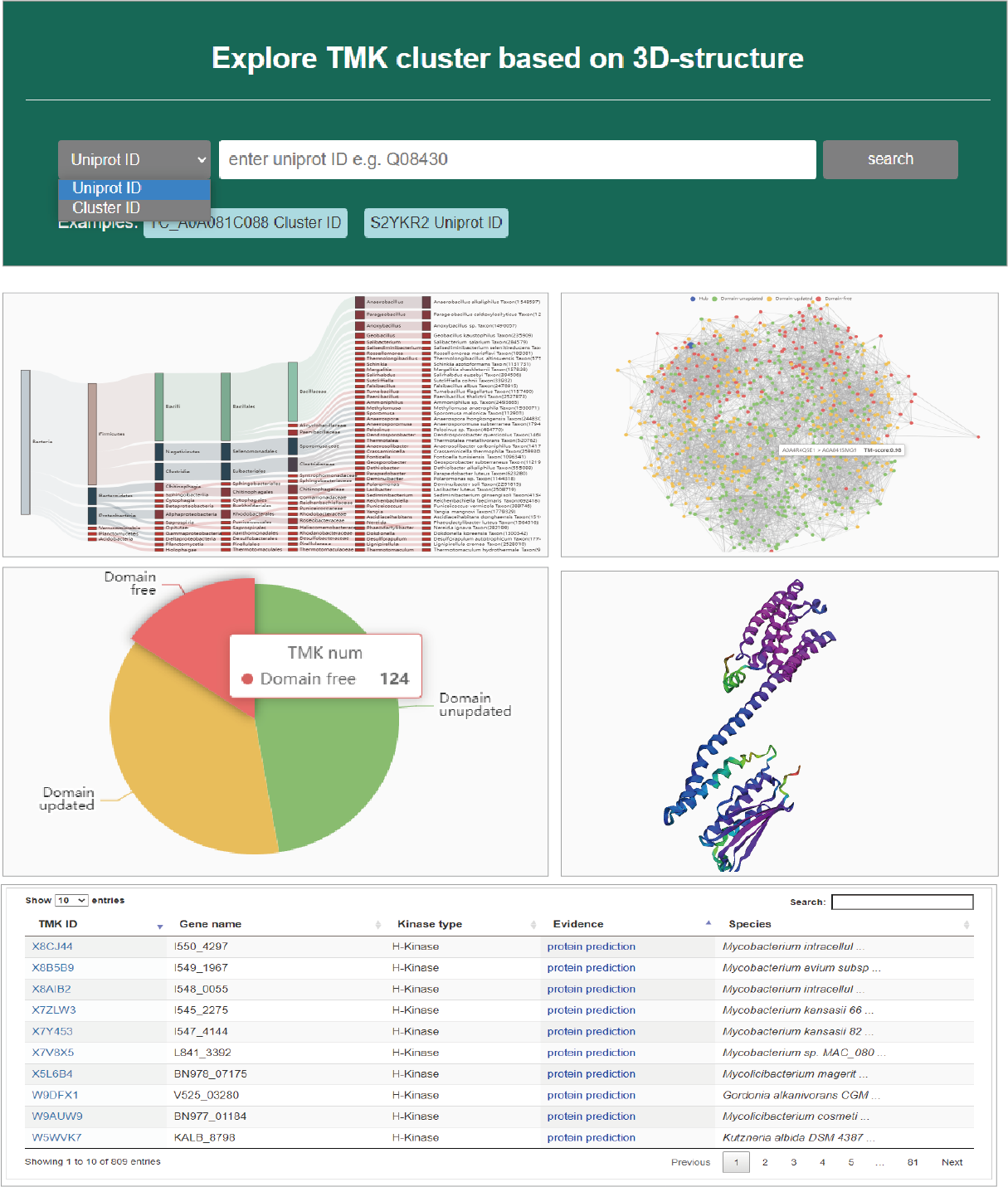
Hide
Query by 3d-structure
TMKingdom also offers a query function by 3D-structure. If users upload a PDB file or offer a uniprot ID corresponding to PDB file, TMKingdom can return hit clusters in an interactive table. In addition, the alignments based on sequence and 3D-strucure can be further displayed below the result table.
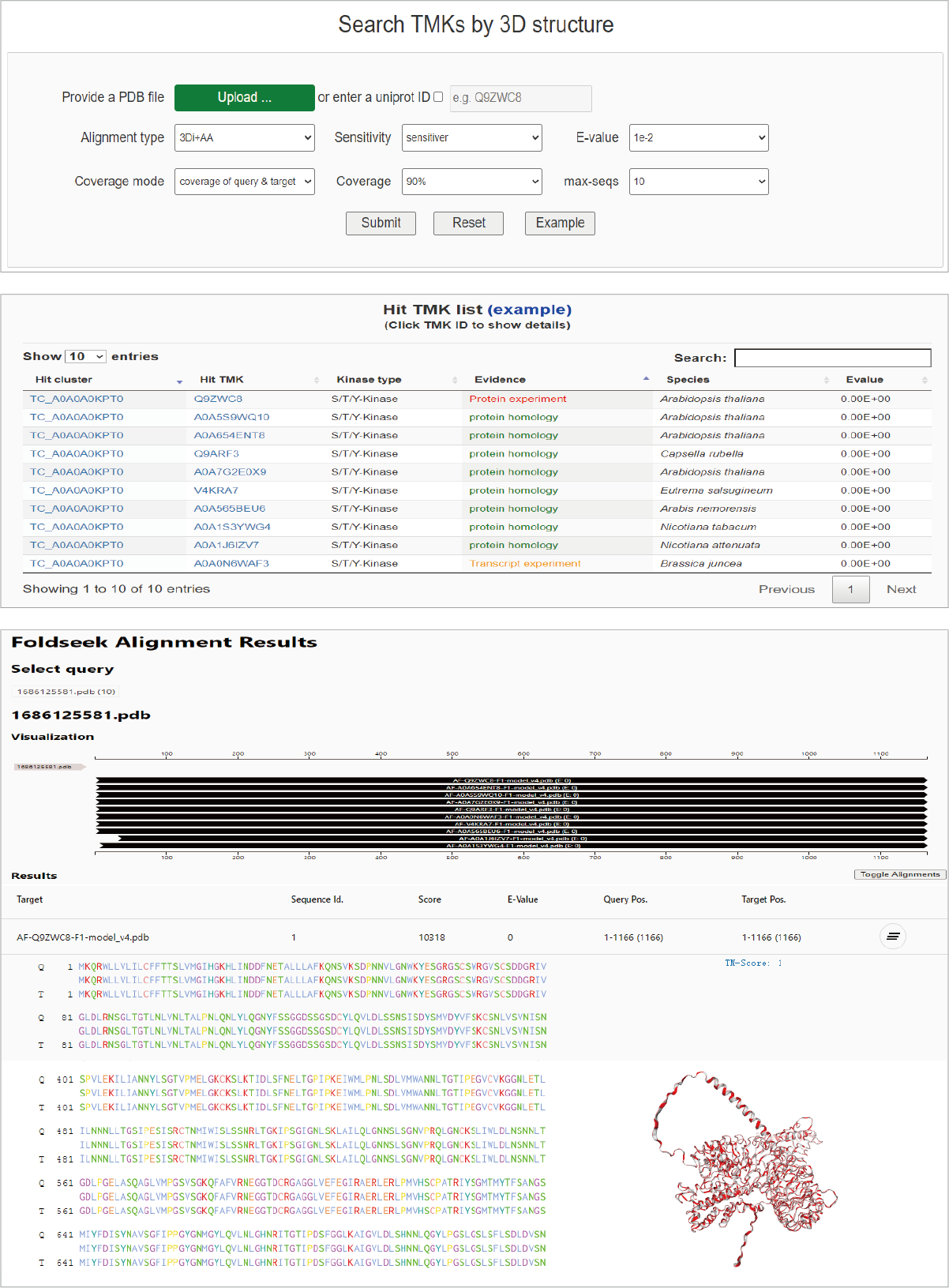
Hide
Summary of cluster
Users can obtain the summary of clusters through the cluster menu. First, a column chart exhibits the distribution of cluster size. Second, a pie chart displays percentages of domain updated by 3D-structure similarity. Users can specify cluster size and updated percentage to list concerned clusters. An interactive table shows cluster size, domain updated status, species number covered.
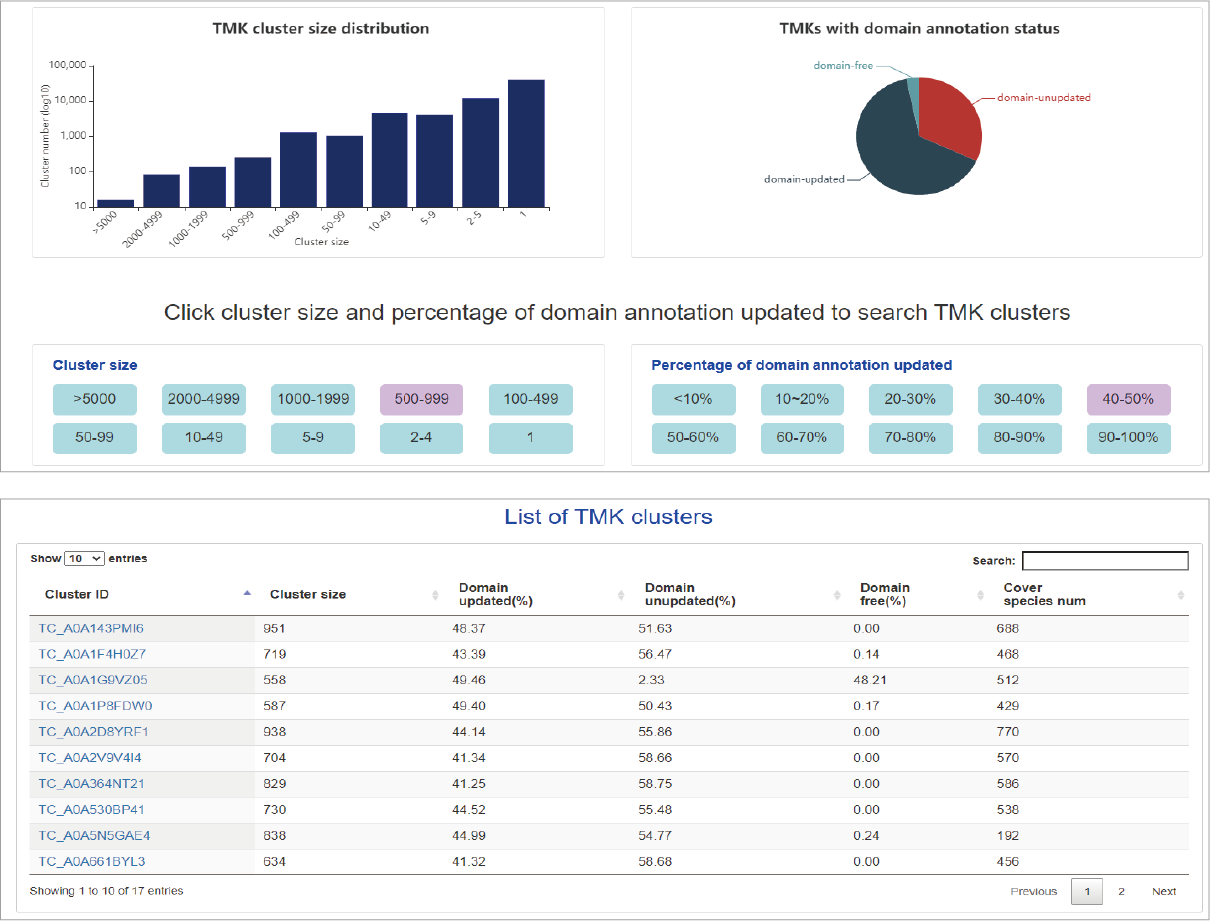
Hide
Download
Download TMKs by species
Through the download menu, users can open the download page to specify a position in species lineage to list concerned species. The TMK sequence and annotation files of each species can be freely downloaded.
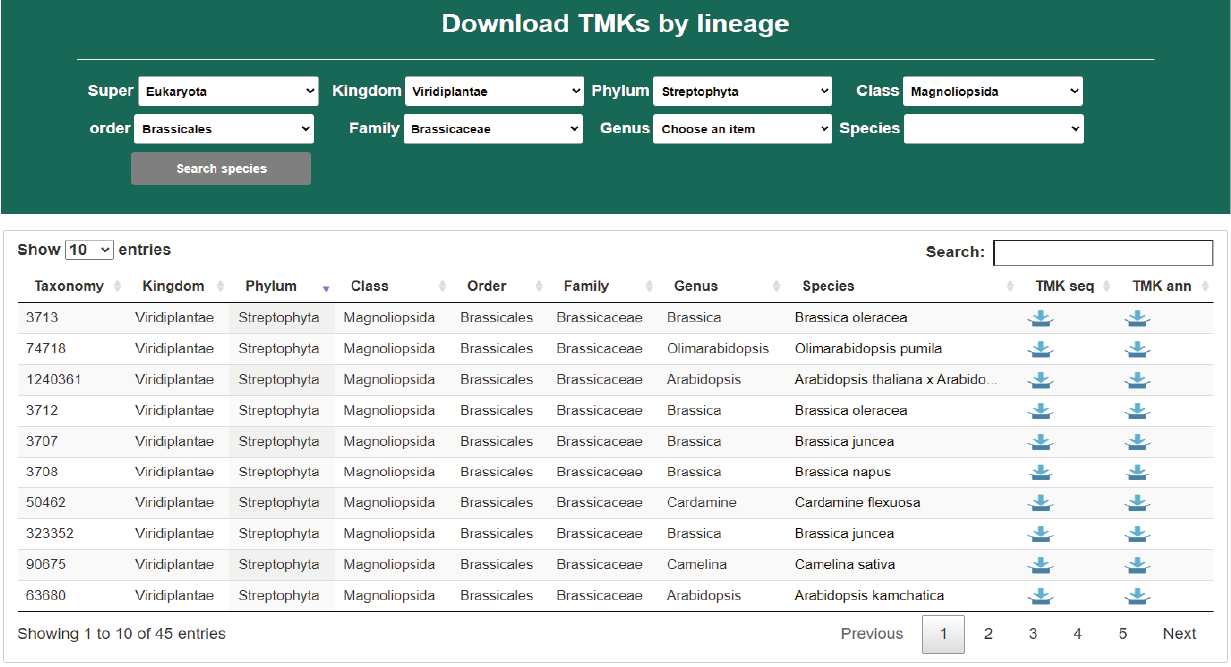
Hide
Contact
Jinding Liu
liujd@njau.edu.cn
References
TMKingdom: a comprehensive resource of massive transmembrane kinases crossing the life kingdom. bioRxiv, 2023.
Cross-kingdom analyses of transmembrane protein kinases show the functional diversity and distinct origins in protists. bioRxiv, 2023.